Understanding amyloid aggregation by statistical analysis of atomic force microscopy images
Adamcik J., Jung JM., Flakowski J., De Los Rios P., Dietler G., Mezzenga R., Nature Nanotechnology 5, 423 (2010).
Abstract
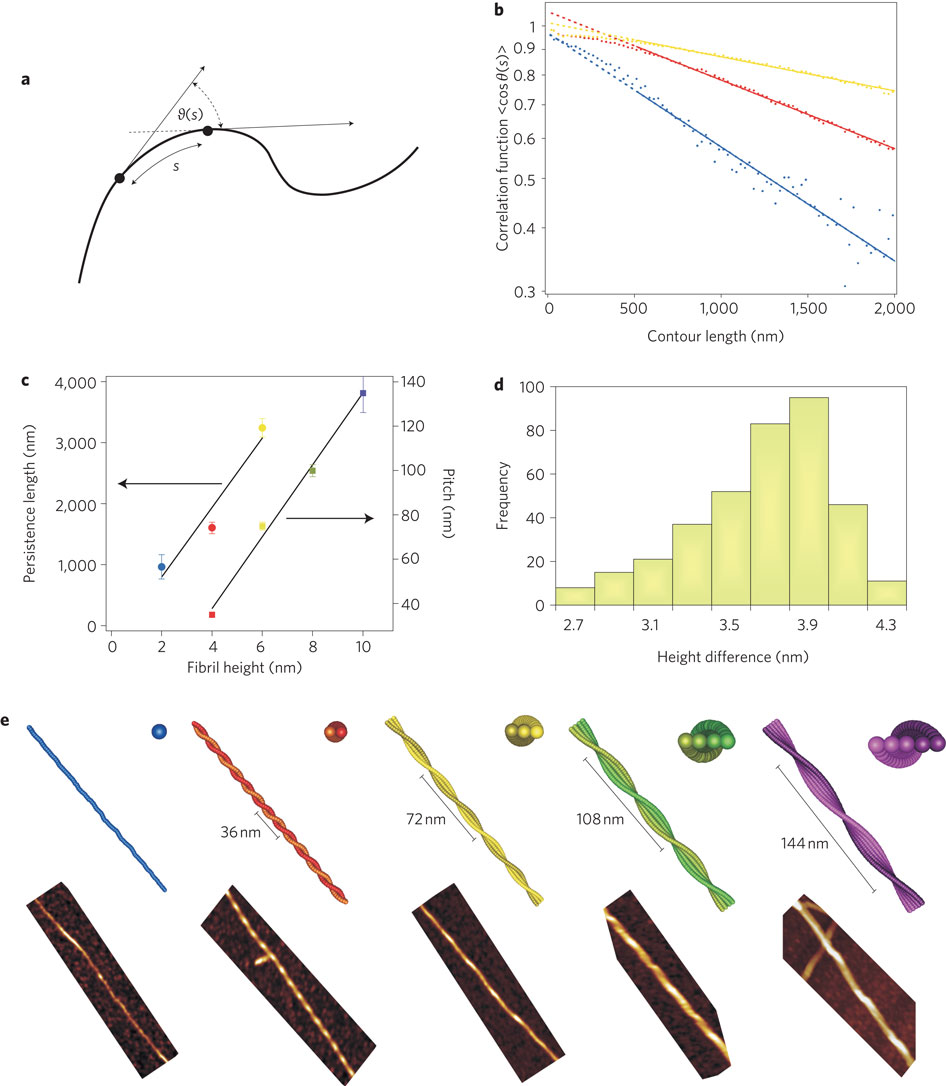
The aggregation of proteins is central to many aspects of daily life, including food processing, blood coagulation, eye cataract formation disease and prion-related neurodegenerative infections1, 2, 3, 4, 5. However, the physical mechanisms responsible for amyloidosis—the irreversible fibril formation of various proteins that is linked to disorders such as Alzheimer's, Creutzfeldt–Jakob and Huntington's diseases—have not yet been fully elucidated6, 7, 8, 9. Here, we show that different stages of amyloid aggregation can be examined by performing a statistical polymer physics analysis of single-molecule atomic force microscopy images of heat-denatured β-lactoglobulin fibrils. The atomic force microscopy analysis, supported by theoretical arguments, reveals that the fibrils have a multistranded helical shape with twisted ribbon-like structures. Our results also indicate a possible general model for amyloid fibril assembly and illustrate the potential of this approach for investigating fibrillar systems.